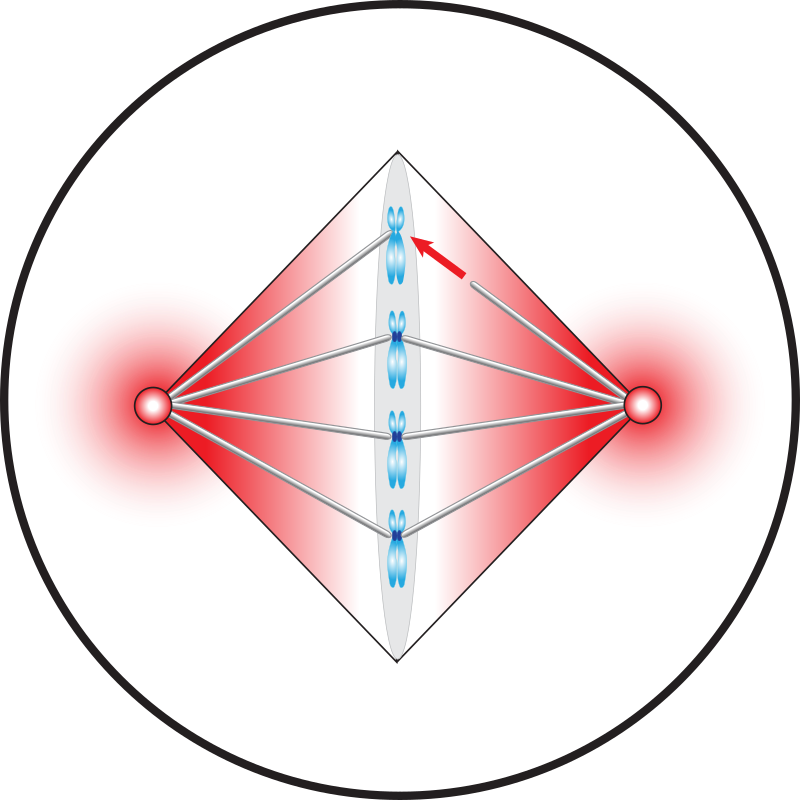
Dynamics of mitotic spindle assembly
Faithful chromosome segregation during mitosis relies on an intricately orchestrated process of mitotic spindle assembly. Spindle assembly involves multimodal coupling between spatiotemporal, mechanical, and biochemical dynamics of hundreds of molecules and macromolecular structures, e.g., microtubules, molecular motors, and chromosomes. We build biophysical models to understand which components and processes are essential and how they function, in assembly, maintenance, and surveillance of the mitotic spindle.
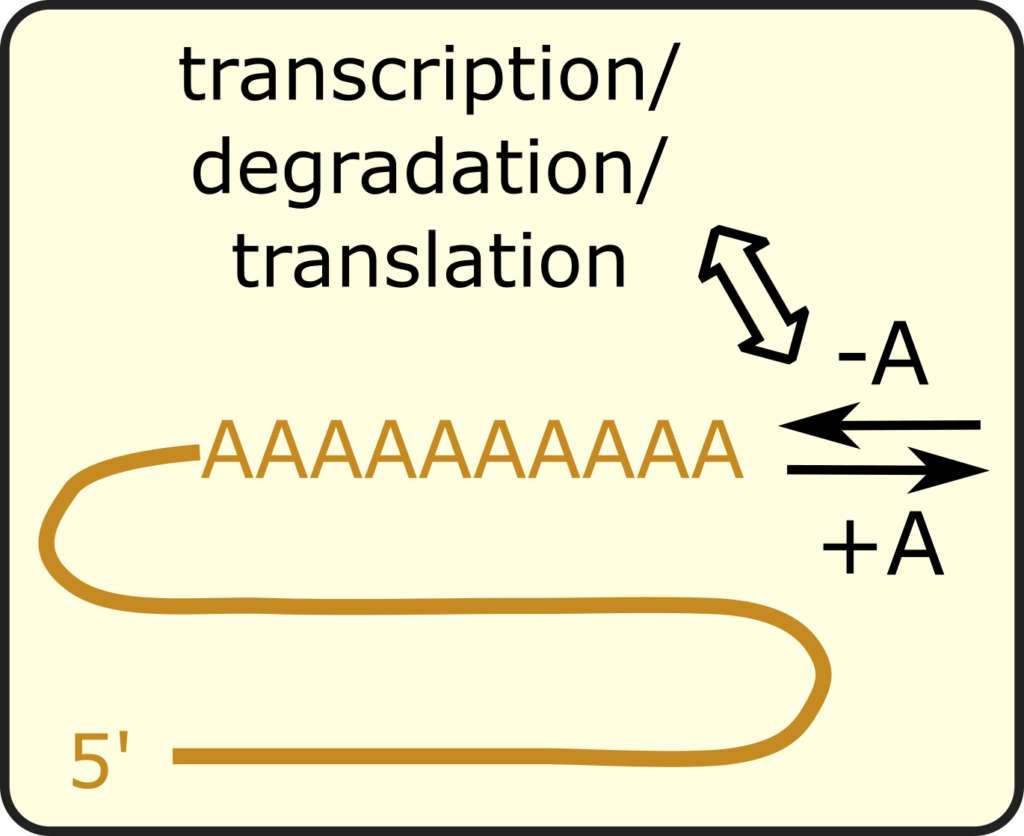
Dynamics of mRNA poly(A) tail
The poly(A) tail is a universal feature for nearly all eukaryotic mRNAs. The molecular machineries that add or remove the poly(A) tail physically interact with the molecular machineries controlling mRNAs throughout their lives, including those mediating transcription, mRNA decay, translation, etc. Hence, poly(A) regulation organizes a highly interconnected system of gene expression control. We are interested in modeling the dynamic regulation of the mRNA poly(A) tail and theorizing about its potential roles in gene expression control.
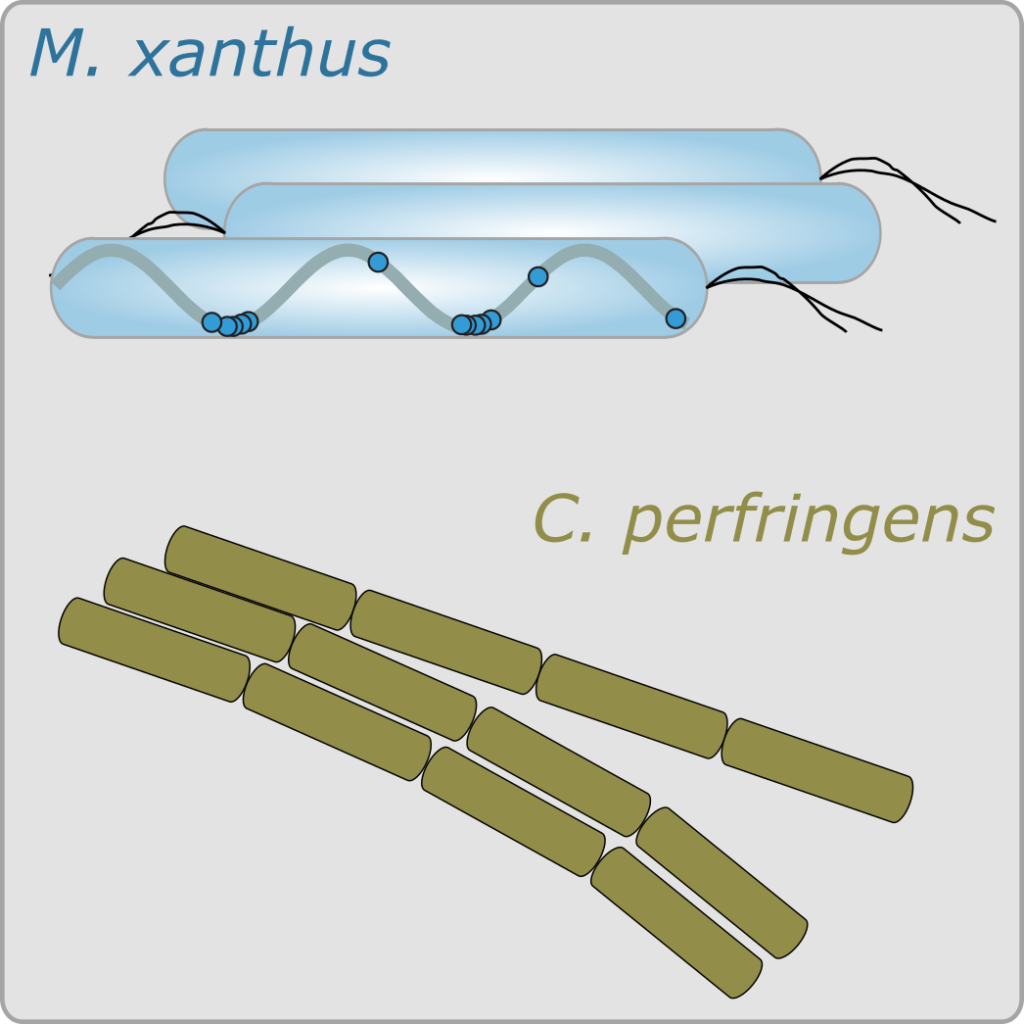
Bacterial gliding motility and control
Controlled, active motility is crucial for survival, virulence, intercellular interactions, and biofilm formation in bacteria. By its nature, the mechanisms driving and regulating bacterial motility involve complex interplay between mechanical, spatiotemporal, and biochemical processes in the bacterial cell. Currently we focus on the gliding motility and motility control in Myxococcus xanthus and Clostridium perfringens.
Spatial co-propagation of phages and host bacteria
We recently constructed a spatiotemporal model to explain the experimentally observed lysis pattern that emerges when a bacterial population encounters phages while expanding in space. Through analyzing the condition and implications of such lysis pattern using the model, we discovered that extended phage-bacteria co-propagation requires an intrinsic physiological balance between phages and bacteria.
